Jiang Y, Langenberg K, Borgdorff V, Duaska M, Post R, Bartulos O, Doornbos M, Braam S, Reijerkerk A, Rasche U.
Eppendorf Application Note, 2019
Abstract
Effective drug discovery and development relies in large part on the availability of predictive preclinical model systems. Application of human cellular models from tissues which are difficult to access, such as cardiomyocytes and neurons, is still challenging. Technologies based on human induced pluripotent stem cells (hiPSC) hold great promise to overcome this challenge. Their routine use in industrial drug research requires a constant supply of stem cell-derived cells of consistent high quality. Researchers from Ncardia® developed a bioprocess for the large-scale manufacturing of cardiomyocytes derived from a variety of healthy and diseased hiPSC lines for implementation into their DiscoverHIT platform.
They expanded hiPSCs as cell aggregates, in a DASbox® Mini Bioreactor System. Aggregate size, hiPSCmarker expression during the expansion phase, and differentiation to the desired cell type were reproducible in three batches. In a proof of concept study, the researchers scaled-up the process using a BioFlo® 320 bioprocess control system. The cells retained key iPSC markers during the expansion phase, providing confidence that stirred-tank bioreactors are suitable to scale-up the production of hiPSC-derived cellular models. The production of several billion cardiomyocytes per batch will enable the screening of large libraries of compounds against phenotypic cellular models representing human biology.
Introduction
Human stem cell technologies enable an end-to-end solution for efficient high-throughput phenotypic screening in drug discovery and development. State-of-the-art hiPSC technologies allow the generation of relevant human cell models for diseases derived from virtually any patient. HiPSC-based cellular assays for drug research need to be robust and reliable, and compatible with automated high throughput screening (HTS) platforms. To mass produce such assays, hiPSCs need to be expanded and differentiated into the desired cell type in a reproducible and scalable manner.

Stem cell characteristics and differentiation are influenced by the culture conditions, as defined by the medium composition, physiochemical environment, physical forces, cell-cell and cell-matrix interactions, and others. The ability to control and reproduce culture conditions is therefore critical to obtaining a standardized cell population. Cultivation in bioreactors opens up new possibilities for process monitoring and control compared with conventional cell culture flasks. pH, temperature, and DO, among other parameters, can be monitored and controlled in real time, making it easier, on the one hand, to achieve conditions resembling the physiological situation, and on the other hand, to reproduce cultivation conditions from batch to batch.
Transfer of cell-based assays from the development phase to commercial production requires producing more cells with the same characteristics. When using cell culture plates or flasks, increasing the amount of cells usually involves “scaleout”, meaning increasing the number of culture vessels. Stirred-tank bioreactors also allow for “scale-up”, meaning increasing the culture volume by using larger vessels. Stirred-tank design allows similar vessel geometries and capabilities at different scales, facilitating a smooth transition to larger volumes.
To enable robust and scalable production of cardiomyocytes for cardiovascular high-throughput screening applications, Ncardia has implemented a bioprocessing pipeline comprising state-of-the-art bioreactor systems which allows them to optimize processes at small scale, to validate the most promising conditions at mid scale of 100 – 250 mL, and to bring the process to the necessary 1 – 10 L scale. The resulting process provides a relatively mature cardiomyocyte model, recapitulating a human cardiomyocyte’s contractile and electrophysiological profile with high predictivity in cardiac toxicity and efficacy assays.
With the current results, Ncardia demonstrates robust large-scale manufacturing of functional human cardiomyocytes derived from a diverse set of (disease) hiPSC lines.
Material and Methods
HiPSC lines
Throughout the study, researchers at Ncardia used their standard hiPSC line for commercial manufacturing of cardiomyocytes, a patient-derived line with a homozygous mutation in KCNQ1 (potassium voltage-gated channel subfamily Q member 1), a CRISPR/Cas9-corrected version of this line, and a patient-derived line with a mutation in MYH7 (myosin heavy chain 7).
Bioprocess systems
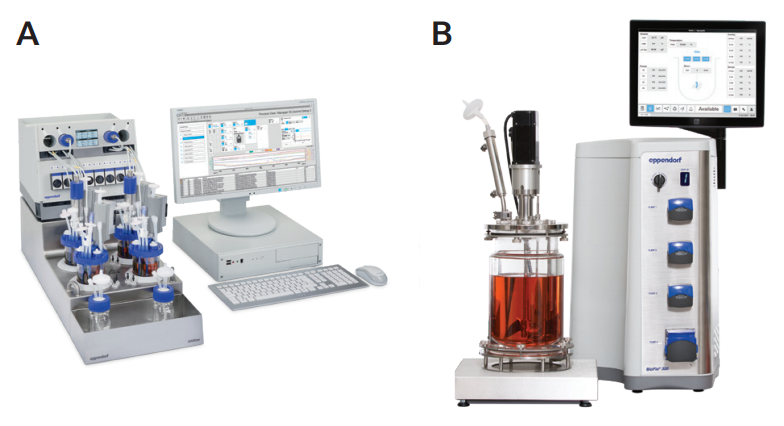
Researchers at Ncardia used differently-scaled bioreactors for process development and scale-up. They identified and optimized critical process parameters, including hiPSC medium, inoculation density, stirring speed, dissolved oxygen, and pH, at 15 mL scale using an ambr® 15 bioreactor (Sartorius®, Germany). In the next step they validated the most promising conditions at small scale (250 mL) using a DASbox Mini Bioreactor System (Eppendorf, Germany; Figure 1A). For manufacturing at the 1 to 10 L scale, they used a BioFlo 320 bioprocess control station (Eppendorf; Figure 1B). The DASbox system and the BioFlo 320 were equipped with BioBLU® c Single-Use Vessels (Eppendorf).
HiPSC expansion and cardiomyocyte differentiation
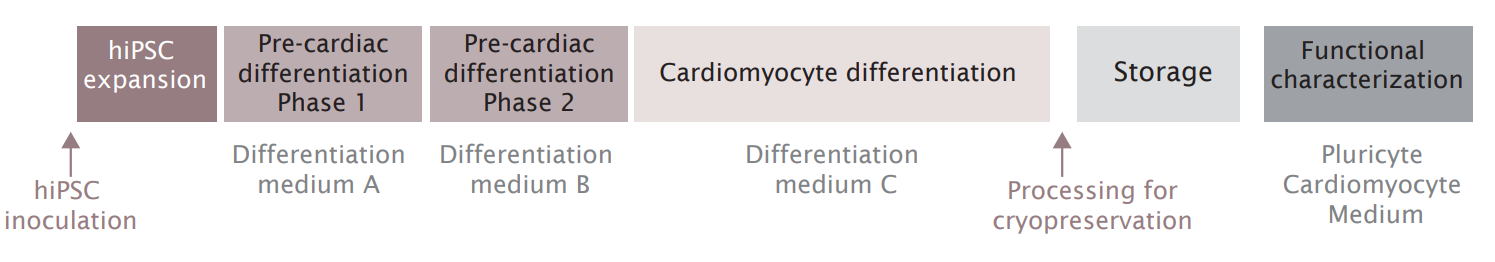
Researchers at Ncardia cultivated hiPSCs and hiPSC-derived cells as cell aggregates. They applied their proprietary differentiation methodology for directed differentiation of hiPSC-derived cardiomyocytes. The workflow for hiPSC expansion, cardiomyocyte differentiation, cryopreservation, and functional testing is represented in Figure 2. At all steps, fully defined media free of serum and growth factors were used.
Analysis of marker gene expression
At several time points, 2 mL samples of the culture were taken from the bioreactor. Single cells were obtained by enzymatic dissociation of the cell aggregates. The expression of marker genes was assessed by flow cytometric measurements. The researchers analyzed the expression of the pluripotency marker genes SOX-2, OCT-3/4 and NANOG, the mesoderm marker BrachyuryT, cardiac troponin T (cTNT), and MLC2v.
Functional characterization of bioreactor-derived cardiomyocytes
For functional characterization, cardiomyocytes produced in bioreactors were thawed and cultivated in plates in Pluricyte® Cardiomyocyte Medium for 8 days, according to Ncardia’s standard protocols. To confirm the quality of bioreactor-derived cardiomyocytes, the researchers analyzed basic cardiomyocyte properties. They studied the organization of cTNT and alpha-actinin by immunofluorescence analysis. Electrophysiological properties (including spike amplitude, field potential duration, and beat rate) and the response to a set of model compounds targeting beta-adrenergic receptors and sodium, potassium, and calcium channels of the hERG-type [2] were investigated in multi-electrode arrays (Maestro™ MEA system, Axion BioSystems®, USA).
Scale-up of hiPSC production and differentiation
To further demonstrate the robustness of the hiPSC-derived cardiomyocyte manufacturing process, the researchers scaled it up from the DASbox Mini Bioreactor System, with a working volume range from 100mL to 250 mL, to the BioFlo320 bioprocess control system equipped with a BioBLU 3c Single-Use Vessel (working volume from 1.25 L to 3.75 L). Culture media and bioprocess parameters, like tip speed, pH, temperature and DO setpoints, were translated from small to bench scale without major modifications.
Results
HiPSC expansion in the DASbox Mini Bioreactor System
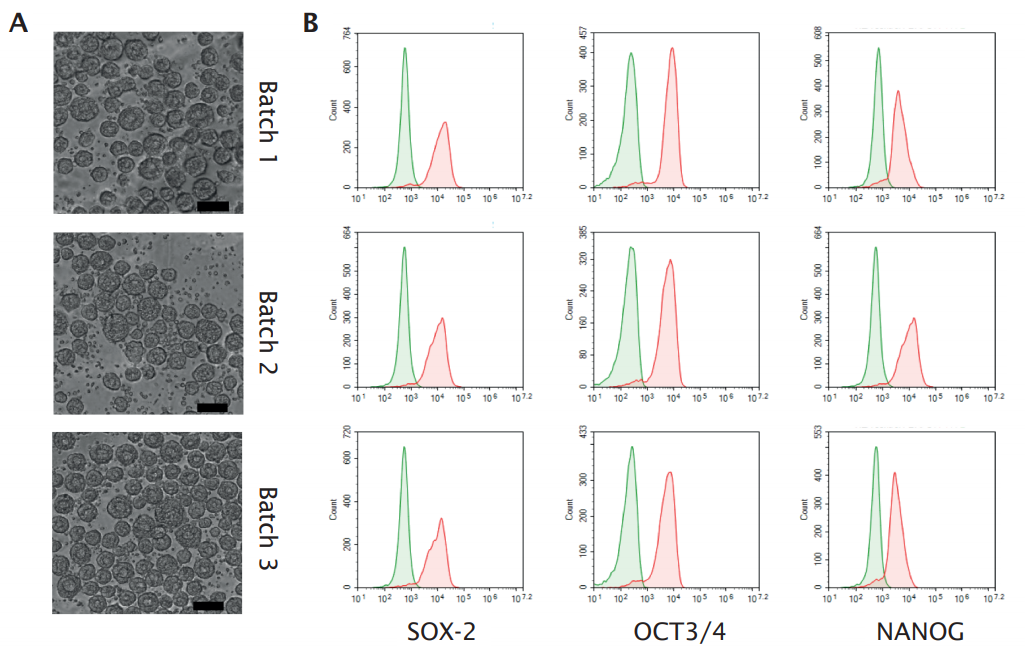
Researchers at Ncardia expanded their hiPSC line for cardiomyocyte manufacturing in a DASbox Mini Bioreactor System equipped with BioBLU Single-Use Vessels. Bright field images of cell aggregates sampled just before start of cardiomyocyte differentiation revealed the formation of spheroids with a diameter of 50 – 100 µm in multiple batches (Figure 3A). More than 90 % of the cells expressed the stem cell markers SOX-2, OCT-3/4, and NANOG (Figure 3B).
d-tank bioreactors
Cardiac differentiation
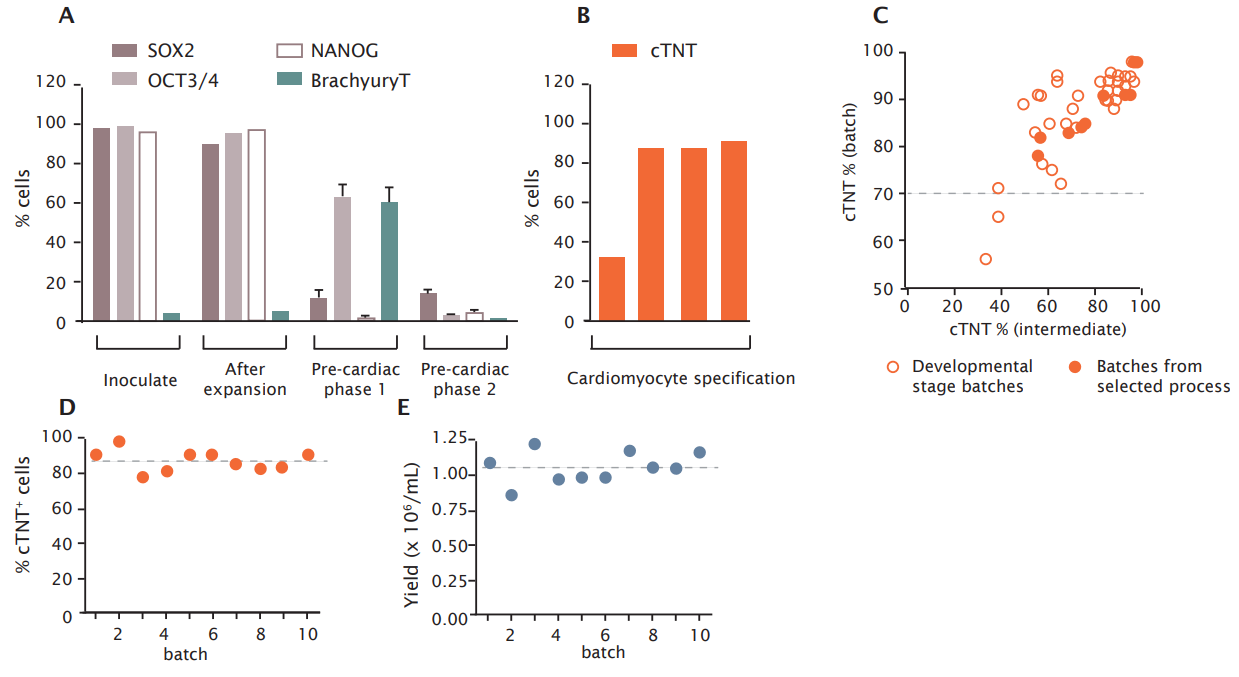
The fate of pluripotency, pre-cardiac, and cardiac markers during the whole differentiation process was assessed in more detail by flow cytometric measurements (Figure 4). At the time of inoculation and after expansion in the bioreactor, high levels of pluripotency markers were observed (Figure 4A). Upon start of differentiation into the pre-cardiac lineage, expression of the stem cell marker SOX-2 rapidly declined and was almost absent after one day of culture. OCT-3/4 was completely diminished after three days from start of differentiation. The mesodermal marker BrachyuryT coincided with pre-cardiac cell development during the first phase of differentiation and was diminished upon directing the cells towards the development of cardiac progenitors and cardiomyocytes. The development of cardiomyocytes was demonstrated by the appearance of the cardiac marker cTNT, reaching maximum levels at the day of harvest (Figure 4B).
Ncardia’s quality control standard is >70 % batch purity with respect to cTNT. Successful cardiomyocyte production meeting this standard was achieved in batches with a cTNT levels of >40 % at an intermediate culture time point (Figure 4C; results are from multiple batches obtained during process development). This result provides a means to abrogate unsuccessful productions already at an early stage. Of note, the two failed batches (intermediate cTNT <40%, final cTNT <70%) were generated with process conditions which were suboptimal compared to the process selected for commercial manufacturing in the future. Thus far, Ncardia’s selected process has resulted in the generation of multiple cardiomyocyte batches with a mean purity of 87.4 ± 5.9 % (Figure 4D) and a mean yield of 1.05 ± 1.11 x 106 /mL (Figure 4E, mean expansion factor was 5.24).
Functional characterization of bioreactor-derived cardiomyocytes
First, the quality of bioreactor-derived cardiomyocytes was confirmed by the presence of beating cardiomyocyte monolayers expressing ventricular-like cardiomyocyte markers (cTNT and MLC2v) after 8 days of culture. 91.7 ± 1.5 % of cultured and dissociated cells from the three batches were cTNT positive (Figure 5A). There was a high level of cTNT+MLC2v+ cells (84 ± 5.6 %, Figure 5B), indicating the maintenance of a pure and ventricular-like cardiomyocyte phenotype. Phase contrast imaging at this time point demonstrated a confluent monolayer of beating cardiomyocytes (Figure 5C). Immunofluorescence analysis of cTNT and alpha-actinin showed a high degree of ultrastructural organization in the cells (Figure 5D).
Second, the quality of the cells in the three batches was corroborated by electrophysiological properties. Bioreactorderived cardiomyocytes showed reproducible field potential signals with pronounced de- and repolarization peaks which allowed accurate assessment of beat rate and field potential duration. The mean beat rate was very regular at 2.1 ± 0.14/s (irregularity in each batch was <2 %), mean field potential duration was 560 ± 47 ms, and mean sodium spike amplitude was 1.7 ± 0.1 mV (Figure 5E-H).
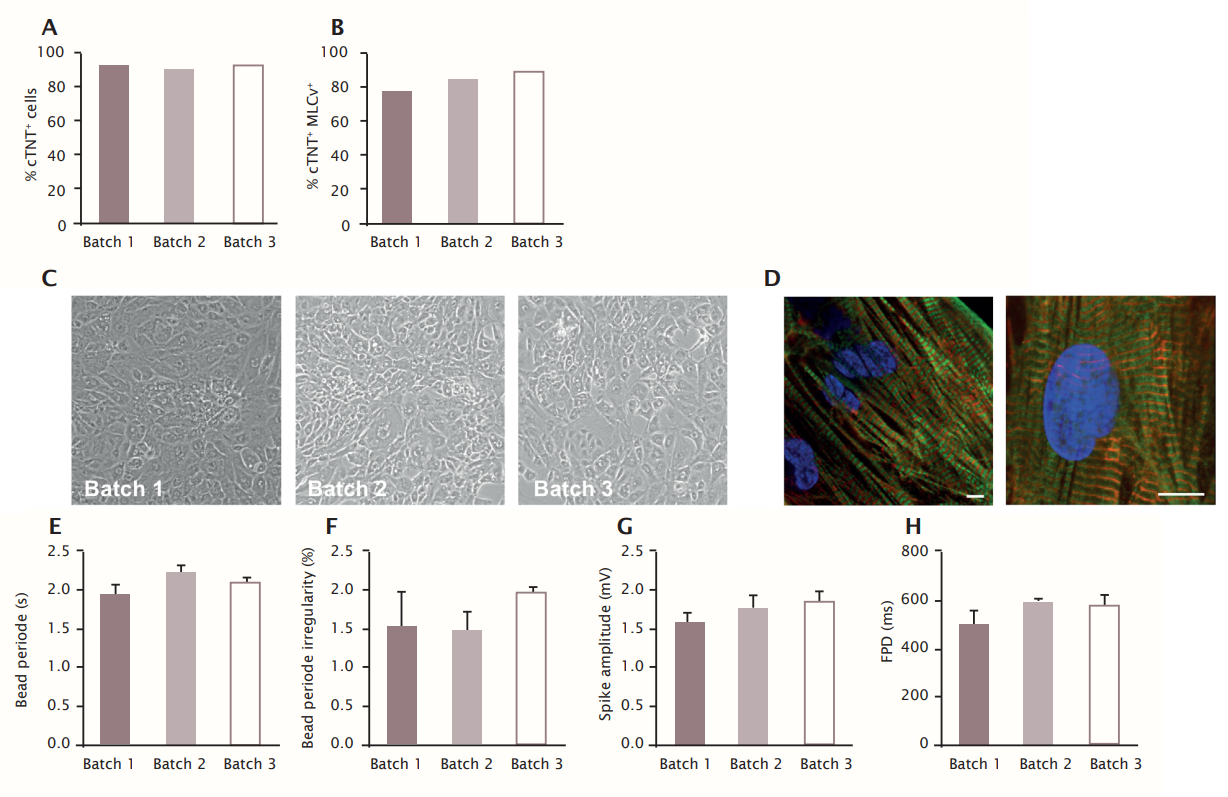
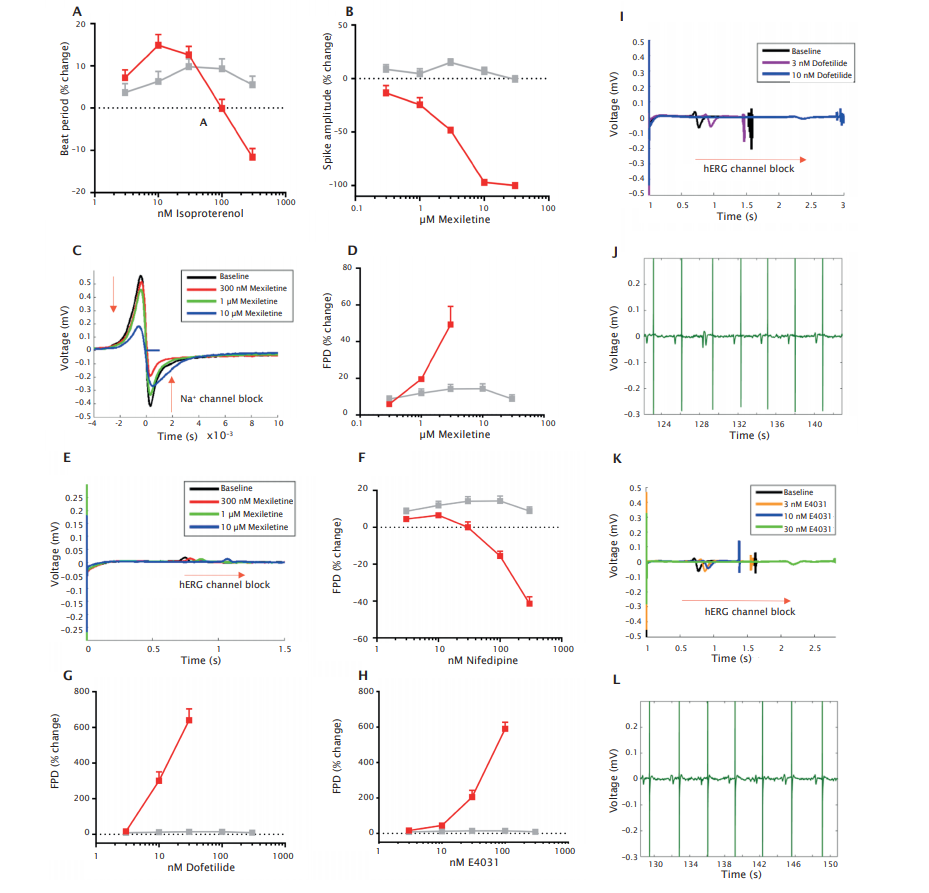
Third, the responses to a set of model compounds which mainly target beta-adrenergic receptors and sodium, calcium and potassium channels of the hERG-type, respectively was assessed. The concentration-dependent response to these cardioactive reference compounds further confirmed the functionality of bioreactor-derived cardiomyocytes. The betaadrenergic receptor agonist isoproterenol enhanced beat rate (Figure 6A), the sodium channel blocker mexiletine reduced sodium spike amplitude (Figure 6B, C) and prolonged field potential duration (FPD) (Figure 6D, E), the Ca2+ channel blocker nifedipine decreased FPD (Figure 6F), hERG channel blockers dofetilide (Figure 6G, I) and E4031 (Figure 6H, K) prolonged FPD and caused TdP-like arrhythmias (Figure 6J, L).
Cultivation of other (disease) hiPSC lines in stirred-tank bioreactors
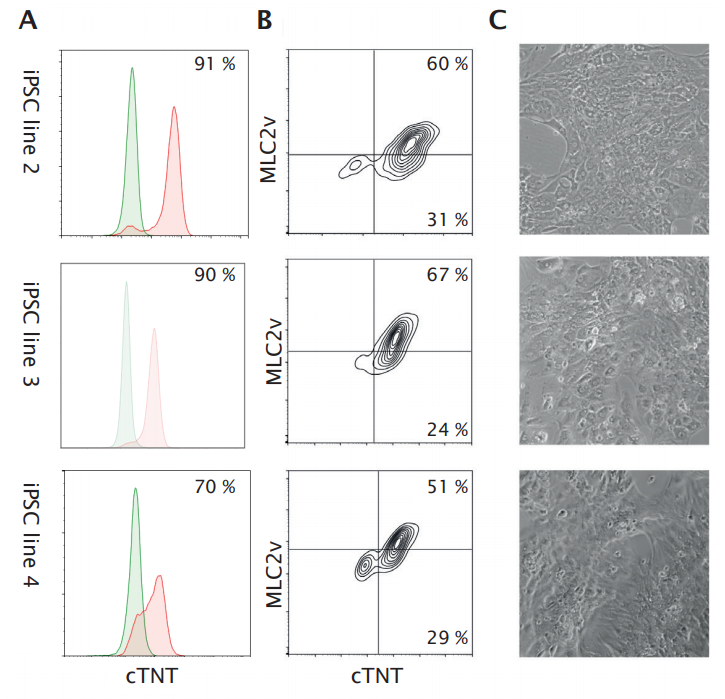
Ncardia’s DiscoverHIT Platform offers the creation of disease models using advanced gene editing technologies. To realize full potential of hiPSC-based disease models in phenotypic screening, broad application of Ncardia’s manufacturing process is a critical component. An efficient workflow requires fast implementation and scale-up of differentiation from any hiPSC line relevant for the disease or application of interest. This can only be done with a robust manufacturing approach. In general, the majority of published protocols for cardiomyocyte differentiation from hiPSCs requires cell line- and cell culture-dependent optimization and can easily lead to heterogeneous differentiation results [1, 2]. To further assess the robustness of their protocol for a bioreactorbased manufacturing process, the researchers tested it for the expansion of a patient-derived line with a homozygous mutation in KCNQ1, a CRISPR/Cas9-corrected version of this line, and a patient-derived line with a mutation in MYH7. The three lines were maintained, inoculated, and differentiated into cardiomyocytes in BioBLU 0.3c Single-Use Vessels using the bioprocess conditions and culture media selected for commercial manufacturing of the standard hiPSC line, without any modification. After differentiation, cardiomyocytes were obtained by enzymatic dissociation of cell aggregates and single cells were cryopreserved using Ncardia’s standard procedure. Cardiomyocyte purity in batches derived from the different hiPSC lines were 91 %, 70 %, and 70 % as assessed by cTNT analysis by flow cytometry (Figure 7A). The quality of bioreactor-derived cardiomyocytes was confirmed by the presence of beating monolayers after 8 days culturing in Pluricyte Cardiomyocyte Medium according to Ncardia’s standard protocols. FACS analysis of these cultured cardiomyocytes obtained from the three hiPSC lines revealed high levels of cTNT+ MLC2v+ cells (Figure 7B), indicating the maintenance of a pure and mainly ventricular-like cardiomyocyte phenotype. Phase contrast imaging at this time point revealed a confluent monolayer of beating cardiomyocytes (Figure 7C).
Bioprocess scale-up
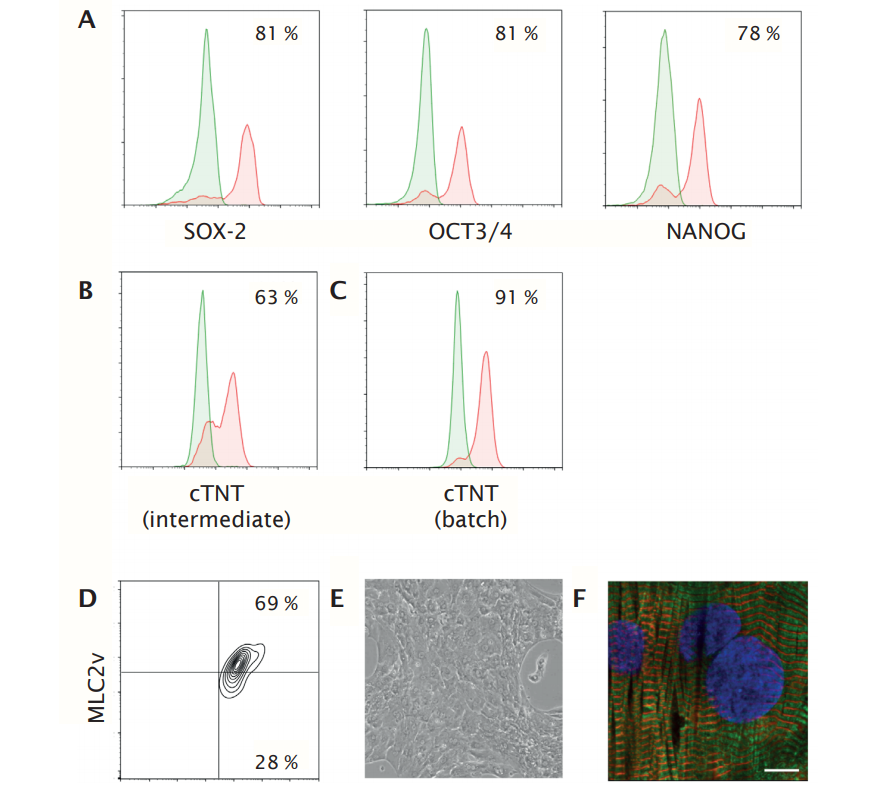
To further demonstrate the robustness of the hiPSC-derived cardiomyocyte manufacturing process, the researchers scaled-up the expansion and differentiation of a selected hiPSC line from a BioBLU 0.3 c Single-Use Vessel (working volume 100 – 250 mL) to a BioBLU 3c Single-Use Vessel (working volume 1.25 L – 3.75 L). In-process assessment of previously identified intermediate critical quality attributes revealed high expression levels of pluripotency marker genes (SOX-2, OCT3/4 and NANOG) before induction of cardiomyocyte differentiation and intermediate cTNT levels of 63 %. Final batch size was 7.5 x109 (2.5 x 106 /mL, conversion rate 12.5) comprising 87 % cTNT+ cells (Figure 8). Further characterization of the cells upon 8 days culture in Pluricyte Cardiomyocyte Medium showed a confluent monolayer of beating cardiomyocytes and high levels of cTNT+ MLC2v+ cells, indicating the maintenance of a pure and ventricular-like cardiomyocyte phenotype (Figure 8D, E). Immunofluorescence analysis of cTNT and alphaactinin showed a high degree of ultra-structural organization in the cells (Figure 8F).
Conclusion
Using a controlled, stirred-tank bioreactor approach Ncardia manufactures functional human cardiomyocytes derived from a diverse set of (disease) hiPSC lines at large scale. This enables the high-quality and large-scale manufacturing of cardiovascular cells required for phenotypic screening of potential drugs in any relevant biological system at a high throughput. Ncardia’s fully controlled bioprocess for manufacturing of hiPSC-derived cardiomyocytes (serum free, growth-factor free differentiation) is highly reproducible in independent batches originating from one hiPSC line. The proprietary process was successfully used, without modifications, for manufacturing of cardiomyocytes from any other hiPSC line tested. With this, the manufacturing process has great potential for the development of a variety of disease-specific models (patient-derived or genetically introduced) for use in Ncardia’s DiscoverHIT Platform. The yield of ~1 x 106 hiPSC-derived cardiomyocytes per milliliter in each bioreactor showed that the process is capable of generating large-scale batch sizes in a robust manner. Ncardia’s bioprocess capabilities allowed easy and fast upscaling to 1010 hiPSC-derived cardiomyocytes per batch, enabling application in high-throughput phenotypic drug discovery campaigns.
This, in combination with a quality-by-design approach, will allow fast process development for manufacturing of other hiPSC-derived cell types and use in DiscoverHIT Platform applications.
Acknowledgement
Ncardia has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 726513.
Literature
- Controlled manufacturing of hiPSC-derived cardiomyocytes in stirred-tank bioreactors enabling highthroughput phenotypic screening. Ncardia Whitepaper. 2019.
- Mulder P. et al. Predicting cardiac safety using human induced pluripotent stem cell-derived cardiomyocytes combined with multi-electrode array (MEA) technology: A conference report. J Pharmacol Toxicol Methods. 91:36-42. May-Jun, 2018.
- Sepac A. et al. Comparison of cardiomyogenic potential among human ESC and iPSC lines. Cell Transpl. 21:2523-2530. 2012
- Sanchez-Freire et al. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J Am Coll Cardiol. 64(5):436-48. Aug 5, 2014


